Contents
Overview
- The procedure details in SeqSphere+ are used to document all laboratory, assembly, and target scan and QC details. In addition an audit trail function (who did what and when) is implemented.
- To control the correct functioning of the NGS machine parameters like throughput (e.g., in GB), Q30, or read length (especially processed) are most valuable.
- There are potentially a number of metrics available to evaluate the quality of WGS data:
- on raw read level - number of reads, number of bases, read length (un-processed and after trimming by quality), average coverage (un-assembled; un-trimmed or trimmed by quality), throughput (related to cluster density on Illumina machines) and
- on assembly level - total sequence length, number of contigs, N50 (or better NG50 that requires the exact genome size, or even better NGA50 that requires the finished genome of the strain under study), average coverage of contigs, percentage of good cgMLST targets.
- However, to control the whole procedure (laboratory & bioinformatics) the percentage of good cgMLST targets (perc. good targets) is most helpful. Good targets fulfill the default Target QC Procedure checks, i.e., same length as reference genes +/- 3 triplets, no ambiguities, and no frame shifts in consensus compared to reference genes. SeqSphere+ shows the perc. good targets value among others (e.g., coverage) and the user can define a threshold for sample success. Low score of perc. good targets usually occur for two reasons:
- most frequently the average coverage for the studied sample is too low or
- an ill-defined ad hoc or (even rarer) stable cgMLST scheme is used.
Effect of Coverage for De Novo Assembling on perc. good targets
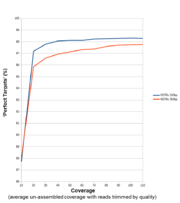
Strains with finished genomes were re-sequenced using Nextera XT library read pairs on a Illumina MiSeq system. Various ‘ad hoc’ schemes were defined with the cgMLST Target Definer and the default Task Template options plus the ‘no variants to ref.-seq.’ option turned on for each of the following listed finished genomes: S. aureus COL (NC_002951; 2.8 MBases genome, 2,391 targets / 77.4% of whole chromosome nucleotides), C. jejuni ATCC 700819 (NC_002163; 1.6 MBases, 1,336 / 80.1%), E. faecium TEX16/DO (NC_017960; 2.7 MBases, 2,340 / 78.3%), E. coli Sakai (NC_002695; 5.5 MBases, 4,333 / 75.1%), and P. aeruginosa PAO1 (NC_002516; 6.2 MBases, 5,266 / 84.6%). With the SeqSphere+ v2.2 pipeline the data were first quality trimmed, downsampled in 10x increments, then de novo assembled (Velvet v1.1.04; automatic mode), and finally the ‘ad hoc’ scheme targets were interrogated for ‘perfect’ matches (good targets with no indel/substitution allowed).
For Illumina data optimal coverage for de novo assembly followed by cgMLST lies around 90-100x. Due to experimental inter-sample variation (≥20%) in Nextera (XT) library preparation and to the trimming of reads, the coverage aimed for in the experimental set-up should be therefore about 130x!
Effect of Coverage for Reference Mapping on perc. good targets
M. tuberculosis strain H37Rv that has a finished genome was re-sequenced using Nextera XT library read pairs on a Illumina MiSeq system. An ‘ad hoc’ scheme was defined from the M. tuberculosis H37Rv genome (NC_000962; 4.4 MBases genome, 3,621 targets / 85% of whole genome nucleotides) with the cgMLST Target Definer and the default Task Template options plus the ‘no variants to ref.-seq.’ option turned on. With the SeqSphere+ v2.2 pipeline the data were first quality trimmed, downsampled in 10x increments, then reference assisted assembled (BWA –sw, v0.6.1-r104; default settings), and finally the ‘ad hoc’ scheme targets were interrogated for ‘perfect’ matches (good targets with no indel/substitution allowed).
For Illumina data optimal coverage for mapping followed by cgMLST lies around 70-80x. Due to experimental inter-sample variation (≥20%) in Nextera (XT) library preparation and to the trimming of reads, the coverage aimed for in the experimental set-up should be therefore about 110x!
Summary
- For de novo assembly it should be aimed in the experimental setup for a slightly higher coverage (130x) than for mapping (110x).
- For de novo assembly the needed coverage is somewhat species-specific.
- The recommended coverage is higher than most ‘SNP’ calling researchers are applying. Trees with similar/identical topology can be achieved also from cgMLST analysis with less coverage.
- The higher coverage is needed for greater reproducibility of results as can be controlled by percentage good/perfect cgMLST targets; i.e., ‘it is known what is unknown’.
- ‘SNP’ researchers frequently do not know where their SNPs are located and never know what is unknown or missed. That is usually no problem for them as they do not aim for continuous surveillance. Their goal is usually ‘just’ one ‘ad hoc’ study or publication.