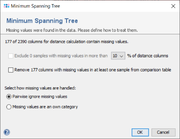
The window allows to select how missing values are handled.
Empty values or values that start with a ? are treated as missing values. There are two ways to exclude (some) missing values from the analysis:
- Exclude Samples with more than 3%, 5%, or 10% missing values in the distance columns
- Samples that have more than the defined missing values are moved to the exclude list.
- Remove columns with missing values in at least one sample from comparison table
- Columns that contain at least one missing value are removed from the comparison table.
If missing values still occur, they can be treated in two different ways:
- Pairwise ignore missing values
- When comparing a non-missing value with a missing value, they are treated as equal. When comparing two missing values, they are also treated as equal.
- Missing values are an own category
- When comparing a non-missing value with a missing value, they are regarded as different. When comparing two missing values, they are treated as equal.
Missing value categories
Three different categories of missing values can appear in SeqSphere+:
- ? (not found): No target sequences were found in the scanning process
- ? (failed): Target QC procedure checks failed (see Target Parameters).
- ? (new): New allele found that have similar (according to the defined parameters) but no identical matches in the allele library. It may be possible to submit the alleles and assign new allele types.
Recommendations
For few targets (e.g., MLVA or MLST):
- use ‘Missing values are own category’ (MLVA, VFDB, or AMRFinder data) OR
- repeat laboratory until there are no more missing values (MLST data).
For many targets and few samples (e.g., WGS data):
- use ‘Remove columns with missing values from distance calculation’ (conservative approach).
For many targets and many samples (e.g., WGS data):
- use ‘Ignore missing values in pairwise comparison’ AND ‘Remove samples with missing values above a definable threshold‘.